Meyer Group
Bioinformatics of RNA Structure
and Transcriptome Regulation
Berlin Institute for Medical Systems Biology
Max Delbrück Centre
Mission
The short- and long-term goals we pursue.
Research
Current areas of research we focus on. Note that we are a purely computational lab .
Computational Methods
Recent computational methods we developed.
Supporters
Funding agencies that support our research.
e-rna web-server
One-stop shop for all of our computational methods and corresponding web-servers.
Press
Our publications and pre-prints, including short summaries.
Recruiting
Information on how to join us as prospective researcher or new collaborator.
Who we are, how to get in touch and directions how to find us .
Our mission
Discover new biological classes of trans RNA-RNA interactions.
Investigate their functional roles, in particular during infection in host-pathogen settings.
Identify targets for therapeutic applications that prevent or enable particular trans RNA-RNA interactions.
Our active areas of research
Regulation of gene expression on RNA level via trans RNA-RNA interactions
Direct trans RNA-RNA interactions between two transcripts can readily influence the expression of transcripts on RNA level. It is only since a few years that we can probe the trans and cis RNA-RNA interactome of cells in a transcriptome-wide manner in a way that rely on a protein-specific interaction. We are thus only starting to explore the functional roles that trans RNA-RNA interactions play in vivo. We have a long-standing interest in trans RNA-RNA interactions and hypothesise that many functional classes of these interactions remain to be discovered, particular in the setting of pathogen-host interactions, see e.g. pdf and pdf. As trans RNA-RNA interactions are comparatively easy to engineer (compared to interactions that require a protein to bind a stretch of RNA), they offer ample potential to be artificially engineered, optimised and/or prevented.
Key results
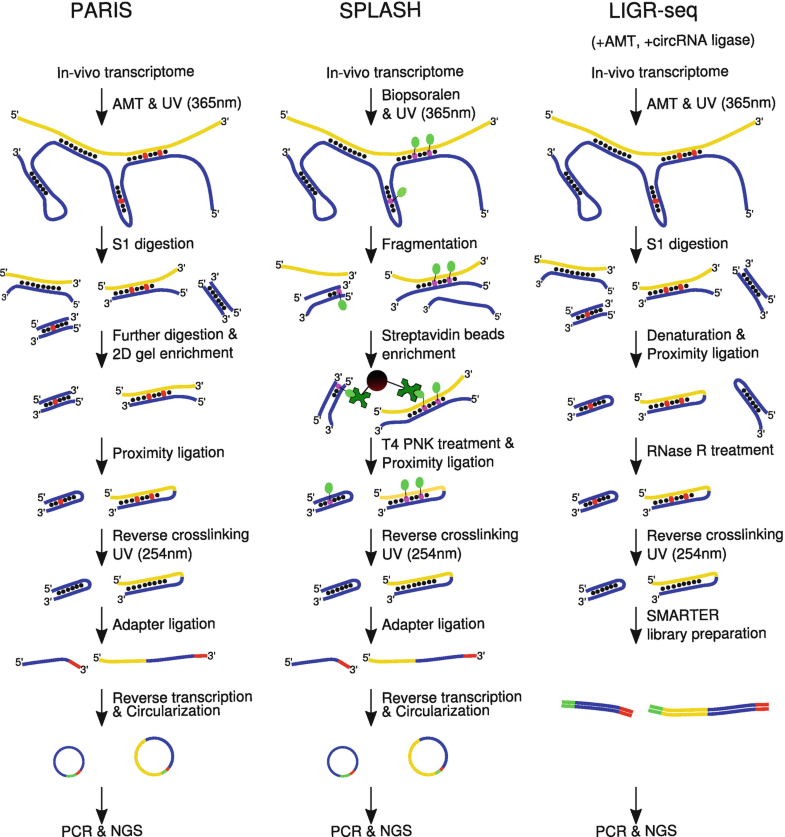
Co-transcriptional RNA structure formation
Transcripts in vivo start folding into transient RNA secondary structure features as soon as transcription starts. This so-called co-transcriptional folding pathway helps the transcript efficiently fold into its functional RNA secondary structure in the cellular environment. Many commonly used computational methods for RNA secondary structure prediction, however, silently assume that the input transcript is (1) already fully synthesized, (2) in thermodynamic equilibrium and (3) that it has no trans interaction partners that may impact its folding. These assumptions generally do not hold for transcripts in vivo.
Key results
- A new computation method CoBold that is capable of detecting transient RNA structure features that may influence the co-transcriptional folding of a given reference transcript and its known reference RNA structure, see pdf, CoBold web-server, CoBold visualization web-server.
- A new computation method CoFold that is capable of capturing one overall effect of co-transcriptional folding into minimum-free energy RNA secondary structure prediction, see pdf and CoFold web-server
- We were the first to annotate several RNA families that have more than one functional RNA structure, see pdf and pdf.
- We have shown that transient RNA secondary structure can be well conserved during evolution, on a level that may be similar to the level of conservation of the final, functional RNA structure, see pdf, pdf and pdf
- We were the first to show that co-transcriptional folding is encoded in the primary sequences of structured RNA genes, see pdf
Regulation of gene expression on RNA level via RNA secondary structures
Transcripts of protein-coding genes are often depicted as linear sticks, e.g. in many text book illustrations of the central dogma of biology. The expression of protein-coding transcripts in vivo, however, may be finely regulated via RNA secondary structure features which are exquisitive sensors of the particular cellular environment. Depending on the cellular environment in vivo, a transcript may respond by expressing one of several potential RNA structures that are all encoded in the primary sequence of the transcript. This is a concept that we have introduced as alternative RNA structure expression, see pdf. We have shown that key processes such as alternative splicing and translation can be regulated by RNA secondary features that are directly encoded into protein-coding transcripts and that regulate these processes in response to the particulars of the in vivo environment.
Key results
- We were the first to show that alternative splicing in the central nervous system and brain can be regulated via local RNA secondary structure features near splice sites which change upon tissue-specific A-to-I editing, see pdf
- We have identified a range of RNA secondary structures that can influence splicing and translation in protein-coding transcripts in a wide range of organisms, for analyses and novel computational methods see pdf, pdf, pdf, pdf, pdf and pdf.
Our computational methods
CoBold
Given an RNA with a known structure, discover transient RNA structure features that may influence the co-transcriptional formation of the reference structure, see pdf, CoBold web-server and CoBold visualization web-server.
CoFold
Given an RNA, predict the minimum-free energy structure while taking the overall effect of co-transcriptional folding into account, see pdf and CoFold web-server.
Transat
Given an RNA and a corresponding multiple-sequence alignment, identify evolutionarily conserved helices (consecutive stretches of base-pairs) including helices from transient, pseudo-knotted and alternative RNA structures, see pdf and Transat web-server.
SimulFold
Given an RNA and a set of evolutionarily related sequences (which are not yet aligned), simultaneously predict the best evolutionarily conserved RNA secondary structure (including pseudo-knots), the best structural multiple sequence alignment as well as the evolutionary tree linking the input sequences, see pdf and SimulFold web-server.
RNA-Decoder
Predict evolutionarily conserved RNA structures that are fully or partly contained in known protein-coding regions. Requires as input a multiple-sequence alignment, see pdf, pdf and RNA-Decoder web-server.
R-Chie
Visualise complex RNA structures and trans RNA-RNA, RNA-DNA and DNA-DNA interactions alongside quantitative data, with evolutionary information (multiple sequence alignments) and in a comparative way, see pdf, pdf and R-Chie web-server (including download of corresponding R code).
BIQ
Search for circular RNAs in transcriptome databases by indexing backsplice junctions, see pdf and BIQ web-server.
e-rna web-server
One-stop shop for all of our web-servers and software.
Our supporters
We thank the following funding agencies for generously supporting our research.




